Nanopore sequencing is a third-generation sequencing method that directly measures long DNA or RNA (Figure 1). The method works by translocating a single DNA strand through a Nanopore in which an electric current signal is measured. The levels of such a signal vary depending on the sequence of the DNA molecule.
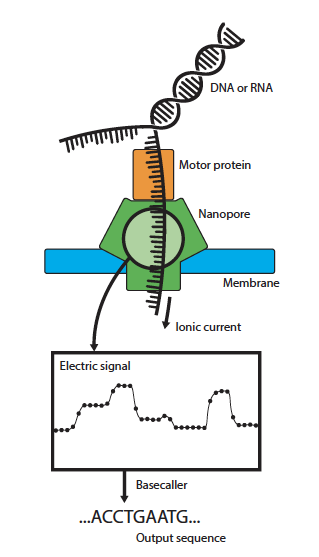
Figure 1. Nanopore sequencing
The relationship between DNA sequence and the electric signal is complex for the following reasons: (1) the electric signal is noisy. (2) The DNA molecules do not translocate through the pore at a constant speed. (3) And the measured signal depends on approximately the 5 bases that are currently inside the pore. Considering only the four canonical DNA bases, this translates to 45 possible signal states.
Translating the electric current to the sequence of DNA bases is called basecalling. State-of-the-art basecalling deep learning models are trained in a supervised manner; meaning that they use signal data from which the DNA sequence is known. However, the amount of annotated Nanopore signal data is scarce and heavily biased towards certain sequenced organisms. These organisms have evolved to have specific DNA sequences in their genome, a bias that can be learned by the basecalling models.
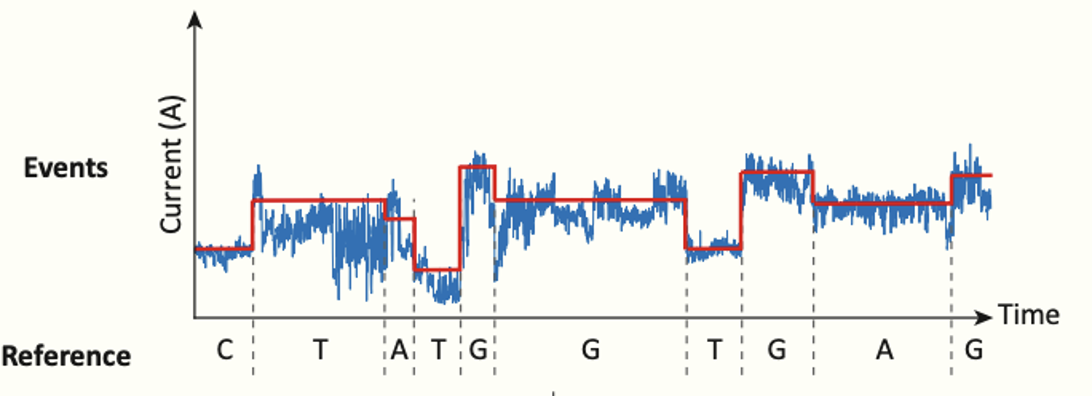
Figure 2. Relationship between DNA sequence and Nanopore raw data. (Adapted from [2])
The aim of this project is to develop a model that can simulate (generate) Nanopore data. Specifically, we would like to develop a model that can capture and reproduce all the variability in Nanopore data given a specific DNA sequence. One direction for such an approach is to use a latent variable model such as a conditional Variational Autoencoder that can produce the high dimensional observation of the electric current time-series given the discrete sequence of nucleotides (i.e. the DNA sequence) (See: ODE-RNN model [1]). Such a model could be trained on a small amount of available annotation and utilize a much larger set of unsupervised data.
Dataset: [available shortly]
Supervisor: Vlado Menkovski, in collaboration with Marc Pagès-Gallego (UMC Utrecht), Jeroen de Ridder (UMC Utrecht)
[1] Rubanova, Yulia, Ricky TQ Chen, and David Duvenaud. “Latent odes for irregularly-sampled time series.” arXiv preprint arXiv:1907.03907 (2019). https://arxiv.org/abs/1907.03907
[2] Yuk Kei Wan, Christopher Hendra, Ploy N. Pratanwanich and Jonathan Göke. “Beyond sequencing: machine learning algorithms extract biology hidden in Nanopore signal data”. Trends in Genetics (2021). https://doi.org/10.1016/j.tig.2021.09.001
